Visualization
Modifies a color by multiplying |
|
Plot a complex matrix with labeling. |
|
Generates a bar chart plot for one or multiple |
|
A collection of |
|
Represents a function that performs a plotting operation either on a |
|
A very generic plotter that compares like estimates across a specified key keyword. |
|
|
Plotter that compares Pauli infidelities from CB, SC, and KNR. |
Very generic plotter that compares like estimates across subsystems of a multi-system register. |
|
Plots estimate fits from IRB. |
|
Plots the estimates of the individual NOX circuits alongside the mitigated estimates. |
|
Plots KNR data as a heatmap. |
|
Very generic plotter that overlays exponential fits onto lightly processed data. |
|
Plots the timestamps of circuit results. |
|
Represents a planar embedding of a quantum device seen as a graph. |
Graph
- class trueq.visualization.Graph(locations, edges=None, show_labels=False, bounds=None, mapping=None)
Represents a planar embedding of a quantum device seen as a graph.
The nodes of the graph represent qubits and the edges are possible couplings. The purpose of this class is to draw this graph in figures.
import trueq as tq import matplotlib.pyplot as plt g = tq.visualization.Graph( {0: (0.1, 0.9), 1: (0.5, 0.9), 2: (0.9, 0.9), 3: (0.5, 0.5), 4: (0.5, 0.1)}, [[0, 1], [1, 2], [1, 3], [3, 4]], ) plt.figure() ax = plt.subplot() cycles = [ tq.Cycle({(0, 1): tq.Gate.cx}), tq.Cycle({(1, 2): tq.Gate.cx}), tq.Cycle({(1, 3): tq.Gate.cx}), tq.Cycle({(3, 4): tq.Gate.cx}), tq.Cycle({(1, 2): tq.Gate.cx, (3, 4): tq.Gate.cx}), ] for idx, cycle in enumerate(cycles): g.add_graph(ax, (0.1 + 0.2 * idx, 1.03), width=0.15, cycle=cycle) # save_figure may not catch things outside of the axis without this plt.tight_layout()
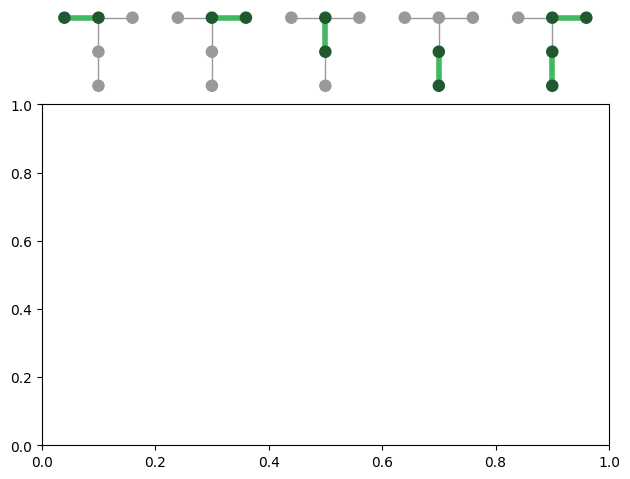
Note
There are a number of class variable such as
NODE_SIZEwhich can be altered on a given instance to change the properties of the displayed graph.- Parameters:
locations (
dict) – A dictionary mapping qubit labels to locations. The locations should fall inside the unit square; they are scaled to the appropriate size whenadd_graph()is called. The argumentboundscan be modified if it is more convenient to specify them in another box.edges (
Iterable) – A list of length-2 tuples specifying the allowed edges.show_labels (
bool|str) – Whether to display qubit label numbers (True), their mapped values based onmapping("mapped"), or none at all (False) on the qubit nodes. Their properties can be adjusted by modifying the class variablesLABEL_OFFSET,LABEL_STYLE, andLABEL_REPR.bounds (
tuple) – A tuple(left, right, bottom, top)indicating the drawing area extents of the coordinates inlocations. By default, this is(0, 0, 1, 1).mapping (
dict) – A dictionary specifying how qubit labels can be mapped to some other convention. The values of this dictionary are shown on plotted graphs ifshow_labels=="mapped".
- NODE_COLOR = '#999999'
The color of a node.
- NODE_SIZE = 4
The radius of a node in points.
- ACTIVE_NODE_COLOR = (0.12423529411764711, 0.34635294117647064, 0.1863529411764707)
The color of a node if a given cycle has a single-qubit gate acting on it.
- LABELED_NODE_DIAMETER = 7
The radius of a node if
show_labelsis true.
- EDGE_COLOR = '#999999'
The color of an edge.
- EDGE_WIDTH = 1
The width of an edge in points.
- ACTIVE_EDGE_COLOR = '#42b863'
The color of an edge if a given cycle has a two-qubit gate acting on it.
- ACTIVE_EDGE_FRAC = 1
The width of an edge as a fraction of node radius.
- LABEL_OFFSET = (0, 0)
The offset of a node label in the same units as
locations
- LABEL_REPR
alias of
str
- LABEL_STYLE = {'color': 'w', 'fontsize': 11, 'ha': 'center', 'va': 'center', 'weight': 'bold'}
Text style properties to draw labels with.
- property edges
A tuple of edges contained in this
Graph.import trueq as tq tq.visualization.Graph.linear(5).edges
(frozenset({3, 4}), frozenset({0, 1}), frozenset({2, 3}), frozenset({1, 2}))- Type:
tuple
- property labels
The sorted system labels in this
Graph.import trueq as tq tq.visualization.Graph.linear(5).edges
(frozenset({3, 4}), frozenset({0, 1}), frozenset({2, 3}), frozenset({1, 2}))- Type:
tuple
- aspen_11()
Instantiates a new
Graphwith a 38 qubit topology found on the Rigetti Aspen-11 chip.import trueq as tq tq.visualization.Graph.aspen_11(show_labels=True).plot()
- Parameters:
show_labels (
bool) – Whether to display qubit label numbers on the qubit nodes.- Return type:
- aspen_m_1()
Instantiates a new
Graphwith an 80 qubit topology found on the Rigetti Aspen-M-1 chip.import trueq as tq g = tq.visualization.Graph.aspen_m_1(show_labels=True) g.LABEL_STYLE["color"] = "midnightblue" g.plot()
- Parameters:
show_labels (
bool) – Whether to display qubit label numbers on the qubit nodes.- Return type:
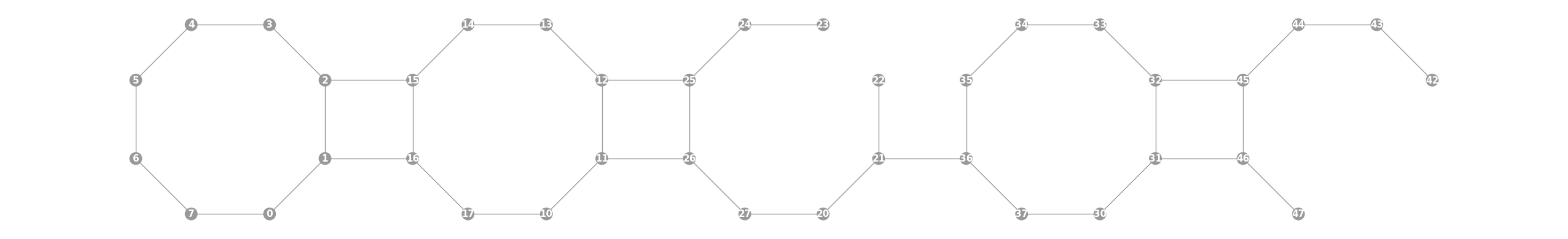
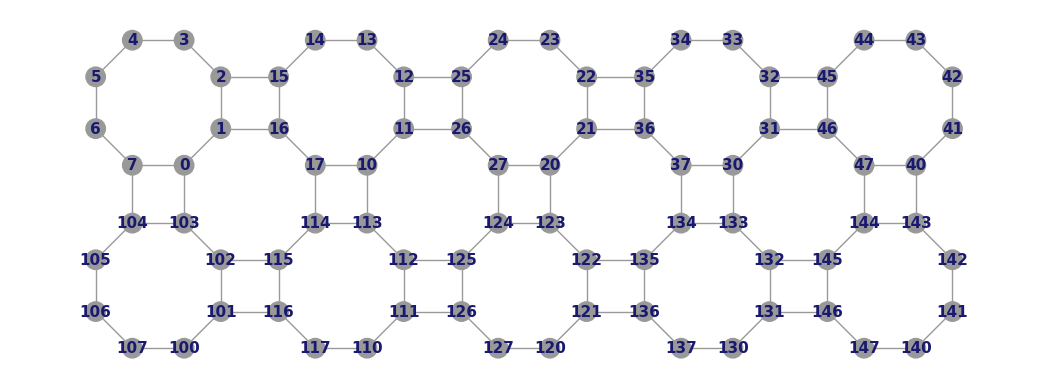
- rigetti_octagon(n_cols, show_labels=False, omit=None)
Instantiates a new
Graphwhere qubits are placed on an_rows-by-n_colsgrid of octagons, where each octagon of eight qubits is connected to each of its neighbouring octagons with two edges.import trueq as tq g = tq.visualization.Graph.rigetti_octagon(1, 2, show_labels=True) g.LABEL_STYLE["color"] = "midnightblue" g.plot()
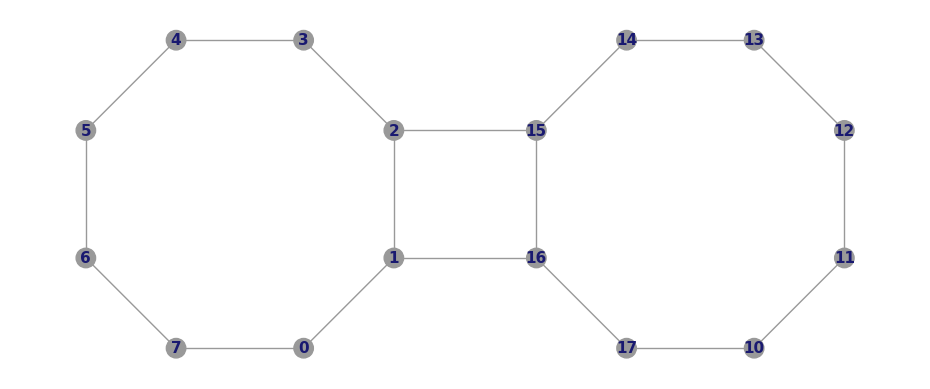
- Parameters:
n_rows (
int) – The number of rows of octagons.n_cols (
int) – The number of columns of octagons.show_labels (
bool) – Whether to display qubit label numbers on the qubit nodes.omit (
int|Iterable) – An iterable of labels or edges to omit from the graph. For example,(2, 3, 5)will omit qubits 2, 3, and 5 from the graph along with incident edges, whereas(2, (3, 5))will omit qubit 2 as well as edge(3, 5)although qubits 3 and 5 will still be present.
- Return type:
- static ibmq_grid(rows, row_major=True, show_labels=False)
Convenience constructor for qubits that lie on a sparse grid of a form particularly popular with IBMQ devices. Each element of
rowsshould be a length-4 list with entries(n_qubits, offset, links, n_int)wheren_qubitsis the number of qubits in the row, all connected as a chain.offsetis the integer horizontal offset of the row from 0.linksis a list of positions in the row at which to add vertical links.n_intis the number of extra qubits to add in each vertical link.
import trueq as tq # create topology shared by Paris, Montreal, and Toronto IBMQ chips, # equivalent to calling Graph.ibmq_27() # note that the 0-qubit first row is used to create the top vertical links g = tq.visualization.Graph.ibmq_grid( [(0, 0, [3, 7], 1), (10, 0, [1, 5, 9], 1), (10, 1, [2, 6], 1)], row_major=False, show_labels=True, ) g.plot()
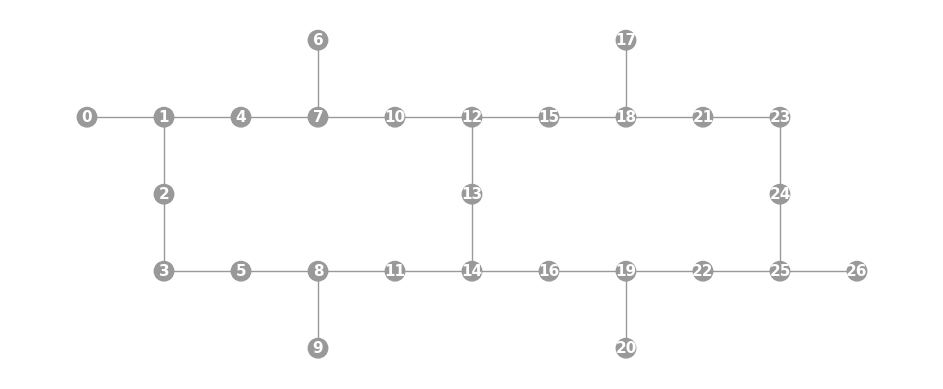
- Parameters:
rows (
list) – A list of lists as described above:row_major (
bool) – Whether qubit labels should increment left-to-right-top-to-bottom (True) or top-to-bottom-left-to-right (False).show_labels (
bool) – Whether to display qubit label numbers on the qubit nodes.
- Return type:
- static ibmq_7(show_labels=False)
Instantiates a new
Graphwith a 7 qubit topology found on some IBMQ devices (including the “Lagos”, “Oslo”, “Nairobi”, and “Perth” chips).import trueq as tq tq.visualization.Graph.ibmq_7(show_labels=True).plot()
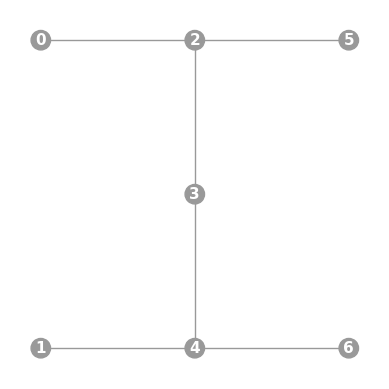
- Parameters:
show_labels (
bool) – Whether to display qubit label numbers on the qubit nodes.- Return type:
- static ibmq_15(show_labels=False)
Instantiates a new
Graphwith a 15 qubit topology found on some IBMQ devices (including the “Melbourne” chip).import trueq as tq tq.visualization.Graph.ibmq_15(show_labels=True).plot()
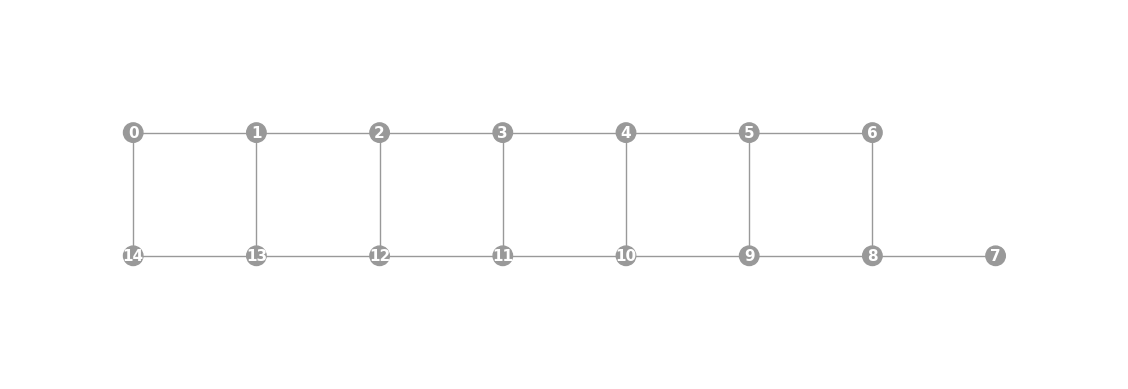
- Parameters:
show_labels (
bool) – Whether to display qubit label numbers on the qubit nodes.- Return type:
- static ibmq_20a(show_labels=False)
Instantiates a new
Graphwith a 20 qubit topology found on some IBMQ devices (including the “Almaden”, “Boeblingen”, and “Singapore” chips).import trueq as tq tq.visualization.Graph.ibmq_20a(show_labels=True).plot()
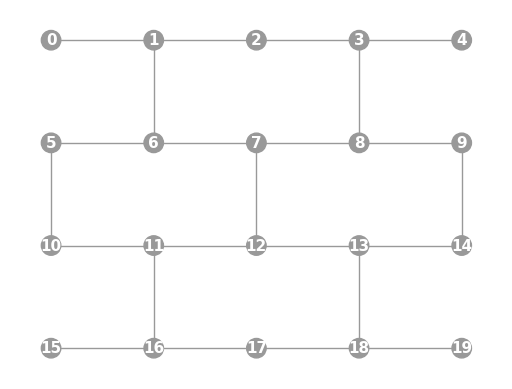
- Parameters:
show_labels (
bool) – Whether to display qubit label numbers on the qubit nodes.- Return type:
- static ibmq_20b(show_labels=False)
Instantiates a new
Graphwith a 20 qubit topology found on some IBMQ devices (including the “Johannesburg” and “Poughkeepsie” chips).import trueq as tq tq.visualization.Graph.ibmq_20b(show_labels=True).plot()
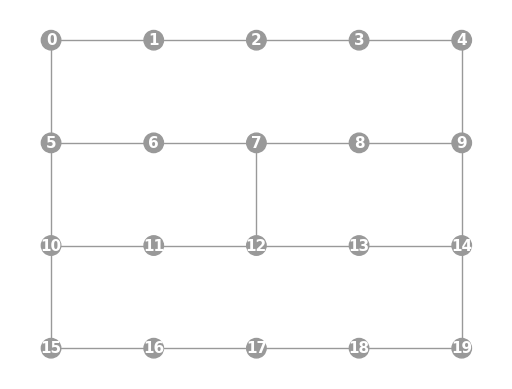
- Parameters:
show_labels (
bool) – Whether to display qubit label numbers on the qubit nodes.- Return type:
- static ibmq_27(show_labels=False)
Instantiates a new
Graphwith a 27 qubit topology found on some IBMQ devices (including the “Montreal”, “Paris”, and “Toronto” chips).import trueq as tq tq.visualization.Graph.ibmq_27(show_labels=True).plot()
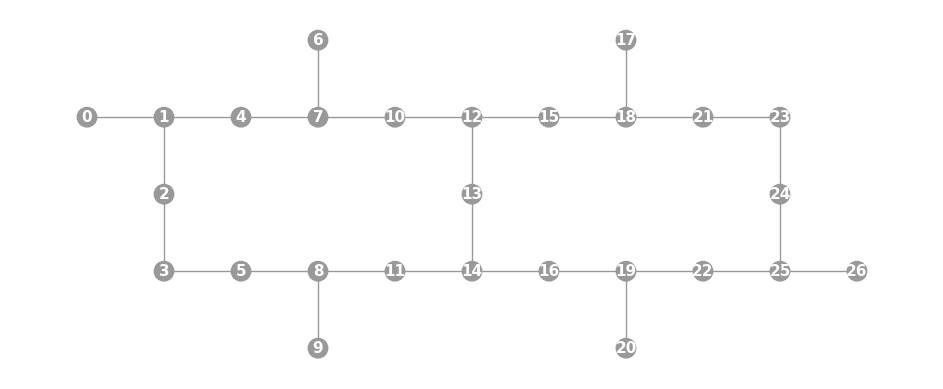
- Parameters:
show_labels (
bool) – Whether to display qubit label numbers on the qubit nodes.- Return type:
- static ibmq_28(show_labels=False)
Instantiates a new
Graphwith a 28 qubit topology found on some IBMQ devices (including the “Cambridge” chip).import trueq as tq tq.visualization.Graph.ibmq_28(show_labels=True).plot()
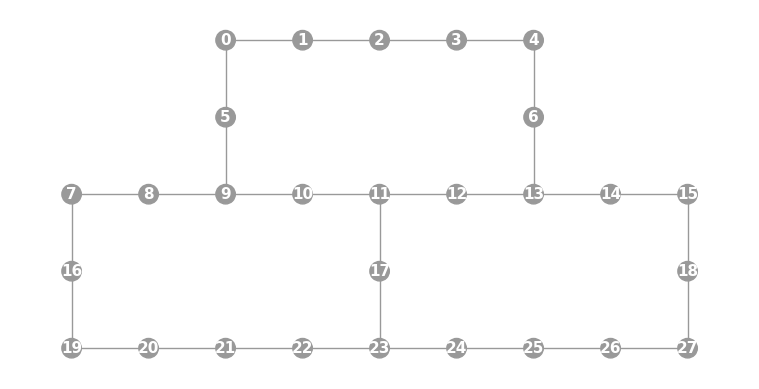
- Parameters:
show_labels (
bool) – Whether to display qubit label numbers on the qubit nodes.- Return type:
- static ibmq_53(show_labels=False)
Instantiates a new
Graphwith a 53 qubit topology found on some IBMQ devices (including the “Rochester” chip).import trueq as tq tq.visualization.Graph.ibmq_53(show_labels=True).plot()
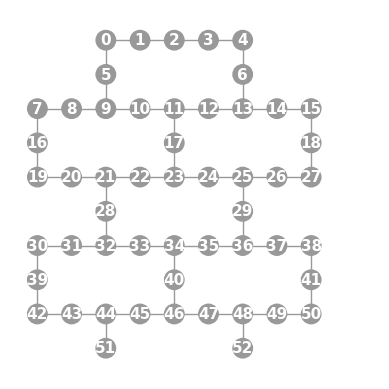
- Parameters:
show_labels (
bool) – Whether to display qubit label numbers on the qubit nodes.- Return type:
- static ibmq_65(show_labels=False)
Instantiates a new
Graphwith a 65 qubit topology found on some IBMQ devices (including the “Manhattan” chip).import trueq as tq tq.visualization.Graph.ibmq_65(show_labels=True).plot()
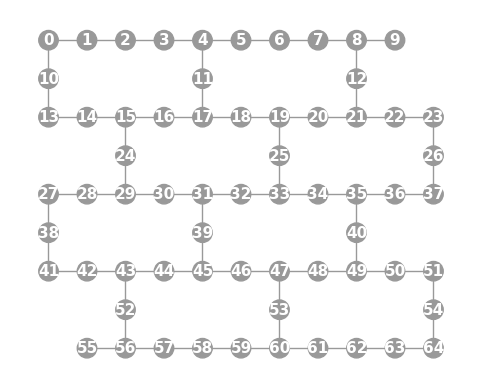
- Parameters:
show_labels (
bool) – Whether to display qubit label numbers on the qubit nodes.- Return type:
- static ibmq_butterfly(show_labels=False)
Instantiates a new
Graphwith a butterfly-shaped 5 qubit topology found on some IBMQ devices (including the “Yorktown” chip).import trueq as tq tq.visualization.Graph.ibmq_butterfly(show_labels=True).plot()
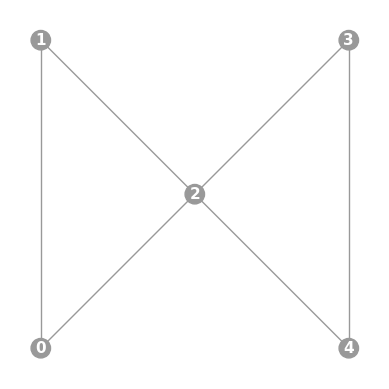
- Parameters:
show_labels (
bool) – Whether to display qubit label numbers on the qubit nodes.- Return type:
- static ibmq_T(show_labels=False)
Instantiates a new
Graphwith a T-shaped 5 qubit topology found on some IBMQ devices (including the “Ourense”, “Vigo”, and “Valencia” chips).import trueq as tq tq.visualization.Graph.ibmq_T(show_labels=True).plot()
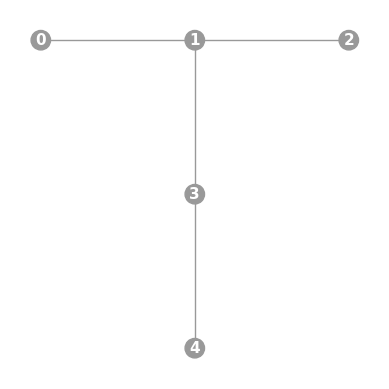
- Parameters:
show_labels (
bool) – Whether to display qubit label numbers on the qubit nodes.- Return type:
- static linear(n, show_labels=False)
Instantiates a new
Graphwith a linear topology.To condense the size of the displayed graph, nodes are placed on a circle.
import trueq as tq tq.visualization.Graph.linear(5, show_labels=True).plot()
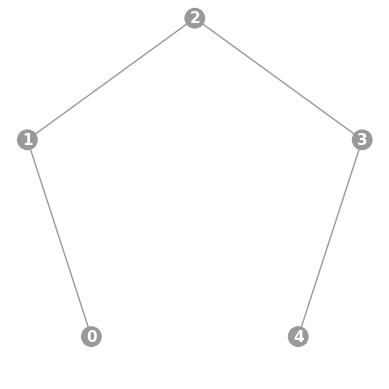
- Parameters:
n (
int|Iterable) – The number of qubits or an iterable of qubit labels.show_labels (
bool) – Whether to display qubit label numbers on the qubit nodes.
- Return type:
- grid(n_cols, show_labels=False)
Instantiates a new
Graphwith a grid topology.Qubit labels start at 0 and are rastered left-to-right from the top-left of the grid.
import trueq as tq tq.visualization.Graph.grid(3, 4, show_labels=True).plot()
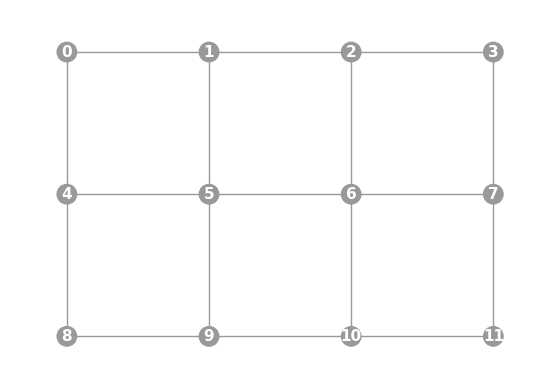
- Parameters:
n_rows (
int) – The number rows in the grid.n_cols (
int) – The number columns in the grid.show_labels (
bool|bool) – Whether to display qubit label numbers on the qubit nodes (True), the labeled coordinate ("mapped"), or no label (False).
- Return type:
- static sycamore(n_rows, n_cols, show_labels=False)
Instantiates a new
Graphwith a diagonal grid topology.Qubit labels are ordered by qubit position sorted with row-major ordering, where the origin is taken as the top left of the embedding.
import trueq as tq tq.visualization.Graph.sycamore(7, 6, show_labels=True).plot()
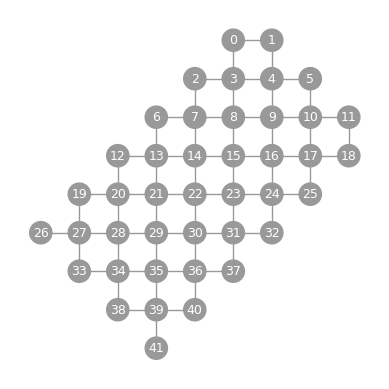
- Parameters:
n_rows (
int) – The number of rows if the grid were rotated 45 degrees clockwise.n_cols (
int) – The number of (zig-zagging) columns if the grid were rotated 45 degrees clockwise.show_labels (
bool|bool) – Whether to display qubit label numbers on the qubit nodes (True), the labeled coordinate ("mapped"), or no label (False).
- Return type:
- to_label(val)
Converts some value to a qubit label.
- Parameters:
val – Some valid value specified in the constructor’s
mappingargument.- Return type:
int- Raises:
KeyError – If
valis unknown.
- from_label(label)
Converts some qubit label to the value specified by the construtor’s
mappingargument.- Parameters:
label (
int) – A qubit label.- Return type:
int- Raises:
KeyError – If
labelis not in this graph.
- add_graph(ax, xy, cycle=None, twirl=None, transform=None, width=None, height=None, xorigin='center', yorigin='bottom', aspect=None)
Embeds the graph into a matplotlib axis at a given position and size.
Certain edges of the graph can be highlighted by passing a cycle.
- Parameters:
ax (
matplotlib.artist.Artist) – The axis (or any other type of artist) to add the graph to.xy (
tuple) – The x and y position at which to add the graph, in coordinates specified bytransform.cycle (
trueq.Cycle) – Coupled edges and active nodes in this cycle will be highlighted in the graph.twirl (
tuple) – Twirled edges and nodes in the twirl will be highlighted in the graph.transform (
matplotlib.transforms.Transform) – Specifies the units ofx,y, width andheight. By default, takes the value blended_transform_factory(ax.transData, ax.transAxes) so that horizontal units are data units, and vertical units are axis units.width (
float) – The width graph, in coordinates specified bytransform. This is optional, seeaspectfor details.height (
float) – The width graph, in coordinates specified bytransform. This is optional, seeaspectfor details.xorigin (
str|float) – One of"left","center", “right”, or any value between 0 and 1. Specifies which point of the graph’s bounding box should be located at position x.yorigin (
str|float) – One of “bottom”, “middle”, “top”, or any value between 0 and 1. Specifies which point of the graph’s bounding box should be located at position `y.aspect (
float) – Force a display aspect ratio of the graph’s bounding box. This is done by computing the largest rectangle of the given aspect ratio that fits in the rectangle of size(width, height). By default, if bothwidthandheightare provided, then these numbers (after being converted to display units) define the aspect ratio. However, if only one ofwidthorheightare given, then a default aspect ratio is defined as the aspect ratio of theboundsset at initialization (default is 1).
- plot(cycle=None, twirl=None)
Plots the graph in its own figure.
- Parameters:
cycle (
trueq.Cycle) – Coupled edges and active nodes in this cycle will be highlighted in the graph.twirl (
tuple) – Twirled edges and nodes in the twirl will be highlighted in the graph.
Multipy Lightness
- trueq.visualization.multiply_lightness(color, amount=0.5)
Modifies a color by multiplying
(1 - lightness)in HSL space by the given amount.Numbers less than 1 result in a lighter color, and numbers greater than 1 result in a darker color.
from trueq.visualization.general import multiply_lightness import matplotlib.pyplot as plt plt.axvline(1, lw=3, color="g") plt.axvline(2, lw=3, color=multiply_lightness("g", 0.3)) plt.axvline(4, lw=3, color="#F034A3") plt.axvline(5, lw=3, color=multiply_lightness("#F034A3", 0.6)) plt.axvline(7, lw=3, color=(0.3, 0.55, 0.1)) plt.axvline(8, lw=3, color=multiply_lightness((0.3, 0.55, 0.1), 1.2))
<matplotlib.lines.Line2D at 0x7f38d92b95e0>
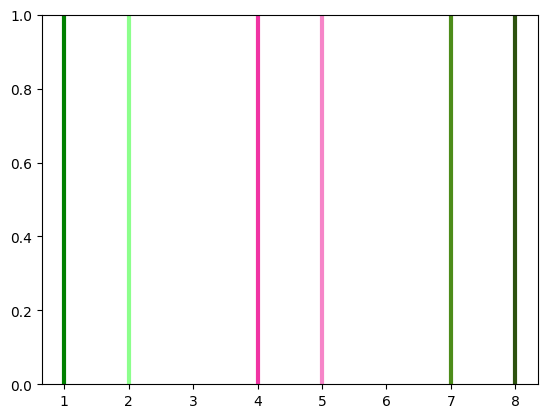
- Parameters:
color (
str|tuple) – A matplotlib color string, hex string, or RGB tuple.amount (
float) – The amount to multiply(1 - lightness)of the input color by.
- Returns:
An RGB tuple.
- Return type:
tuple
Plot Mat
- trueq.visualization.plot_mat(mat, xlabels=None, ylabels=None, show_values=True, abs_max=None, ax=None, xgrid=None, ygrid=None, blocks=None)
Plot a complex matrix with labeling.
The plot will have the following properties:
Color is dictated by the complex angle of each cell.
Transparency (alpha) is decided by the magnitude of each cell.
Size of plot scales based on shape of
mat.
import trueq as tq import numpy as np phase = [np.exp(1j * t * np.pi) for t in np.linspace(0, 1, 8)] tq.plot_mat(np.outer(phase, phase))
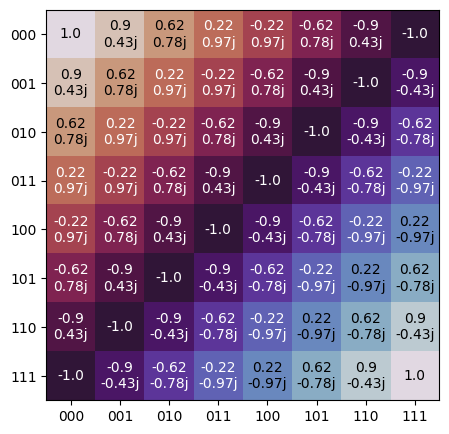
- Parameters:
mat (
numpy.ndarray-like) – A real or complex matrix.xlabels (
int|Iterable|NoneType) – EitherNone, an iterable of labels for the x-axis, or the dimension of the subsystems where the x-axis labels are the computational basis states as inferred from the size ofmat. IfNone, the subsystem dimension will be inferred automatically.ylabels (
int|Iterable|NoneType) – EitherNone, an iterable of labels for the y-axis, or the dimension of the subsystems where the y-axis labels are the computational basis states as inferred from the size ofmat. IfNone, the subsystem dimension will be inferred automatically.show_values (
bool) – Whether to show the numeric values of the matrix in each cell.abs_max (
NoneType|float) – The value to scale absolute values of the matrix by; the value at which plotted colors become fully opaque. By default, this is the largest absolute magnitude of the input matrix.ax (
matplotlib.Axis) – An existing axis to plot on. If not given, a new figure will be created.xgrid (
int|Iterable|NoneType) – An integer specifying the number of columns of separation between vertical grid lines, or a list of integers specifying all of the 0-indexed column numbers before which to place a grid line. IfNone, no vertical gridlines are placed.ygrid (
int|Iterable|NoneType) – An integer specifying the number of rows of separation between horizontal grid lines, or a list of integers specifying all of the 0-indexed row numbers before which to place a grid line. IfNone, no horizontal gridlines are placed.blocks (
Iterable|NoneType) – An iterable of members of the form((i, j), (n_rows, n_cols)), where(i, j)are row and column indices of the matrix specifying the top-left position of a block, and where(n_rows, n_cols)are the number of rows and columns of the block. A rectangular outline will be drawn around each block. IfNone, no blocks will be drawn.
Plot Results
- trueq.visualization.plot_results(*results, labels=None, normalize=None, sparse_cutoff=True, axis=None, style='adjacent', error_bars=False)
Generates a bar chart plot for one or multiple
Resultsobjects using a variety of user-specified plotting styles.Valid
styleinputs are:"adjacent": Results are plotted on the same graph next to each other"overlay": Results are plotted on the same graph on top of each other"separate": Results are plotted on a grid of separate axes
import trueq as tq from trueq.visualization import plot_results r1 = tq.Results({"000": 0.2, "010": 0.1, "110": 0.4, "111": 0.3}) r2 = tq.Results({"001": 0.1, "100": 0.2, "110": 0.3, "111": 0.4}) r3 = tq.Results({"011": 0.3, "010": 0.4, "101": 0.1, "101": 0.2}) plot_results(r1, r2, r3)
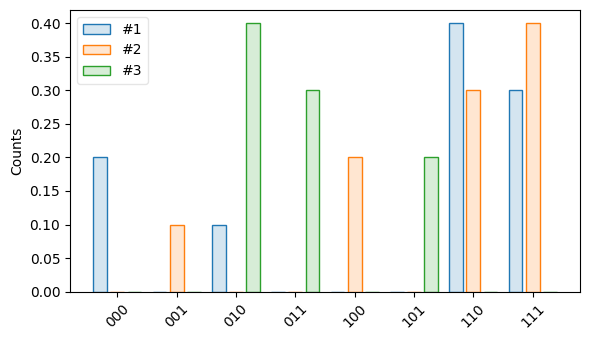
If
error_barsis set toTrue, then each bar on the plot that comes from a results object wheren_shotsis greater than1will have a shot-noise error bar attached to it. The size of these error bars are computed as follows. Suppose the \(i^\text{th}\) bitstring has \(y_i\) counts and comes from results with a total number of shots \(n\). Then we can consider the beta distribution \(\operatorname{Beta}(y_i, n-y_i)\) as the error of that particular bar, and we choose the extents of the error bar as the \(\alpha/2\) and \(1-\alpha/2\) percentiles of this distribution, where the confidence level \(\alpha\) is taken fromget_cl(). The justification is that the Dirichlet distribution \(\operatorname{Dir}(y_1,\ldots,y_N)\) is the posterior distribution of the multinomial data \((y_1,\ldots,y_N)\) under the improper uninformative prior \(\operatorname{Dir}(0,\ldots,0)\), so that \(\operatorname{Beta}(y_i, n-y_i)\) is the posterior marginal distribution of index \(i\). These bounds are asymptotically equal to \(\pm z\sqrt{\frac{y_i}{n}(1-\frac{y_i}{n})/n}\) where \(z\) effects the normal confidence level \(\alpha\).- Parameters:
*results – One or more
Resultsinstances.labels (
Iterable|NoneType) – EitherNoneor a list of string labels corresponding to the inputresults. IfNone, default labels will be applied to the plot.normalize (
NoneType|Bool) – EitherNoneor a boolean value to specify whether results should be normalized for plotting. WhenNone, results appearing on the same axis will all be normalized when their numbers of shots differ.sparse_cutoff (
Bool) – Boolean value to specify whether to clip small values.axis (
NoneType|matplotlib.pyplot.axis|Iterable) – EitherNoneor a single or multiple matplotlib axis instances on which to plot the results.style (
str) – A string to specify the plot style. Valid options are"adjacent","overlay"and"separate".error_bars (
bool) – Whether or not to plot shot-noise errorbars, as described above.
- Raises:
ValueError – If no results were provided or if the results have different values of
n_measurements.
Compare
- class trueq.visualization.plotters.comparisons.compare
A very generic plotter that compares like estimates across a specified key keyword.
Note
This class typically should not be invoked directly by users; the main plotting method is exposed dynamically by the attributes
trueq.CircuitCollection.plotandtrueq.estimate.EstimateCollection.plotif relevant.- static plot(fit, names, keyword, label_rotation=0, axes=None, filename=None)
Plots a comparison of the value of one or more estimates specified by
namesversus the value ofkeywordfound in each estimate’skey. Within each subplot, all other keywords and values are constant.import trueq as tq all_circuits = tq.CircuitCollection() for p in [0.01, 0.02, 0.03, 0.04]: circuits = tq.make_srb(0, [4, 20, 40]) tq.Simulator().add_depolarizing(p).run(circuits) all_circuits.append(circuits.update_keys(p_depolarizing=p)) all_circuits.plot.compare("p", "p_depolarizing")
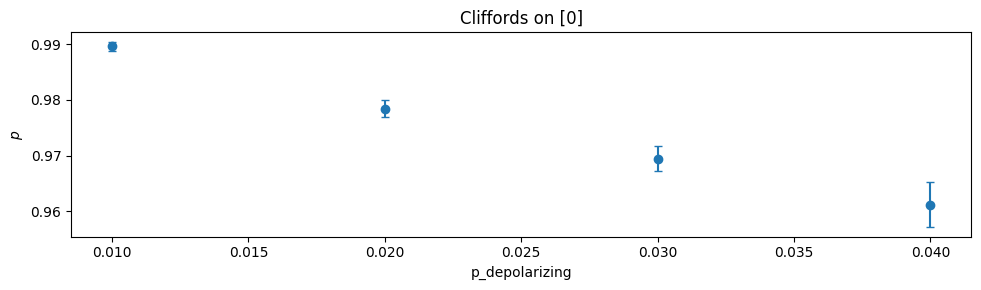
- Parameters:
fit (
EstimateCollection|CircuitCollection) – A collection of circuits or their fit.names (
Iterable|str) – A name or a list of names specifying which estimate parameters should be included in the plot as y-values.keyword (
str) – A keyword found in the estimates’keys whose values the given parameters will be compared against.label_rotation (
float) – The rotation of x-axis labels in degrees.axes (
Iterable) – An iterable of existing axes. If none are provided, new axes are created. Note that axis instances can be repeated in this list if you want to force the combination of subfigures that would normally be distinct, seeaxes()for an example.filename (
str) – An optional filename to save the file as. The extension determines the filetype; common choices are “.png”, “.pdf”, and “.svg”. If no extension is given, PNG is chosen.
Compare Pauli Infidelities
- class trueq.visualization.plotters.comparisons.compare_pauli_infidelities
Plotter that compares Pauli infidelities from CB, SC, and KNR.
Note
This class typically should not be invoked directly by users; the main plotting method is exposed dynamically by the attributes
trueq.CircuitCollection.plotandtrueq.estimate.EstimateCollection.plotif relevant.- static plot(fit, graph=None, axis_size=(8, 3.5), axes=None, filename=None)
Plots a comparison of all Pauli infidelities measured by CB, KNR, or SCexperiments. In the case of SC, the process infidelity of the entire cycle, which is the average of all Pauli infidelities, is overlaid. Every unique experiment and sublabel combination is shown on a separate subplot.
import trueq as tq circuits = tq.make_cb({0: tq.Gate.x, 1: tq.Gate.x}, [2, 8]) sim = tq.Simulator().add_stochastic_pauli(pz=0.04).add_overrotation(0.04) sim.run(circuits) circuits.plot.compare_pauli_infidelities()
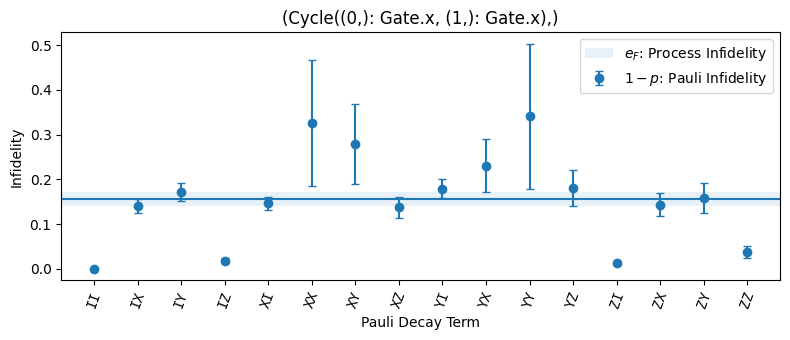
- Parameters:
fit (
EstimateCollection|CircuitCollection) – A collection of circuits or their fit.graph (
Graph) – Optionally,Graphobject to display device topologies to the right of each subplot.axis_size (
tuple) – The width and height of each subplot axis in inches.axes (
Iterable) – An iterable of existing axes. If none are provided, new axes are created. Note that axis instances can be repeated in this list if you want to force the combination of subfigures that would normally be distinct, seeaxes()for an example.filename (
str) – An optional filename to save the file as. The extension determines the filetype; common choices are “.png”, “.pdf”, and “.svg”. If no extension is given, PNG is chosen.
Compare RB
- class trueq.visualization.plotters.comparisons.compare_rb
Compares SRB and XRB infidelities vs twirling groups.
Note
This class typically should not be invoked directly by users; the main plotting method is exposed dynamically by the attributes
trueq.CircuitCollection.plotandtrueq.estimate.EstimateCollection.plotif relevant.- static plot(fit, axes=None, filename=None)
Plots a comparison of process infidelity
e_Fmeasured by SRB and/or stochastic fidelitye_Smeasured by XRB. The comparison is made across twirling groups and is the natural plotting function formake_crosstalk_diagnostics().import trueq as tq circuits = tq.make_crosstalk_diagnostics([0, 1, 2], [4, 32]) tq.Simulator().add_depolarizing(0.05).run(circuits) circuits.plot.compare_rb()
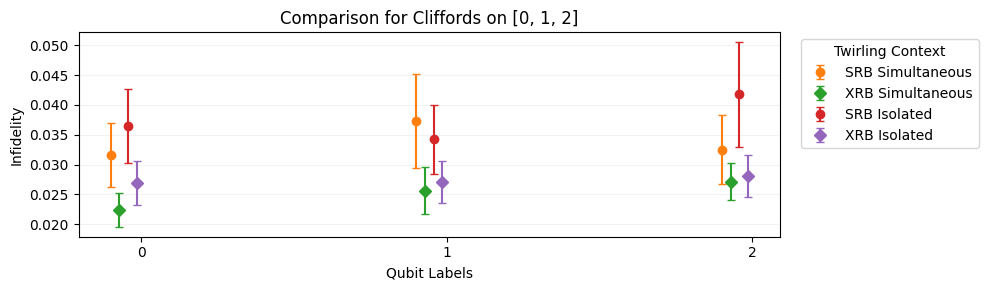
- Parameters:
fit (
EstimateCollection|CircuitCollection) – A collection of circuits or their fit.axes (
Iterable) – An iterable of existing axes. If none are provided, new axes are created. Note that axis instances can be repeated in this list if you want to force the combination of subfigures that would normally be distinct, seeaxes()for an example.filename (
str) – An optional filename to save the file as. The extension determines the filetype; common choices are “.png”, “.pdf”, and “.svg”. If no extension is given, PNG is chosen.
Compare Twirl
- class trueq.visualization.plotters.comparisons.compare_twirl
Very generic plotter that compares like estimates across subsystems of a multi-system register.
Note
This class typically should not be invoked directly by users; the main plotting method is exposed dynamically by the attributes
trueq.CircuitCollection.plotandtrueq.estimate.EstimateCollection.plotif relevant.- static plot(fit, names, axes=None, filename=None)
Plots a comparison of the value of one or more estimates specified by
names. The comparison is made across twirling groups. For example, if some protocol like SRB is performed on qubits in isolation as well as simultaneously, then this function can be used to compare these situations for any of the reported estimates.import trueq as tq circuits = tq.make_crosstalk_diagnostics([0, 1, 2], [4, 32]) tq.Simulator().add_depolarizing(0.05).run(circuits) circuits.plot.compare_twirl("p")
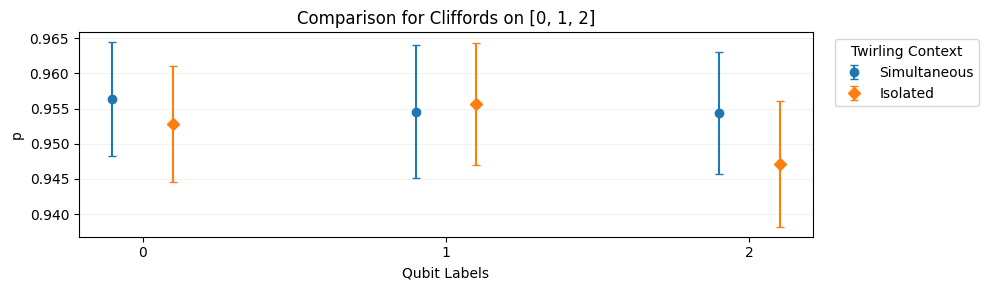
- Parameters:
fit (
EstimateCollection|CircuitCollection) – A collection of circuits or their fit.names (
Iterable|str) – A name or list of names specifying which estimate parameters should be included in the plot.axes (
Iterable) – An iterable of existing axes. If none are provided, new axes are created. Note that axis instances can be repeated in this list if you want to force the combination of subfigures that would normally be distinct, seeaxes()for an example.filename (
str) – An optional filename to save the file as. The extension determines the filetype; common choices are “.png”, “.pdf”, and “.svg”. If no extension is given, PNG is chosen.
IRB Summary
- class trueq.visualization.plotters.irb.irb_summary
Plots estimate fits from IRB.
Note
This class typically should not be invoked directly by users; the main plotting method is exposed dynamically by the attributes
trueq.CircuitCollection.plotandtrueq.estimate.EstimateCollection.plotif relevant.- static plot(fit, axes=None, filename=None)
Plots data from an IRB experiment. Relevant information from associated SRB and XRB experiments is also shown, see the IRB guide for details.
- Parameters:
fit (
EstimateCollection|CircuitCollection) – A collection of circuits or their fit.axes (
Iterable) – An iterable of existing axes. If none are provided, new axes are created. Note that axis instances can be repeated in this list if you want to force the combination of subfigures that would normally be distinct, seeaxes()for an example.filename (
str) – An optional filename to save the file as. The extension determines the filetype; common choices are “.png”, “.pdf”, and “.svg”. If no extension is given, PNG is chosen.
NOX
- class trueq.visualization.plotters.mitigation.raw_nox
Plots the estimates of the individual NOX circuits alongside the mitigated estimates.
Note
This class typically should not be invoked directly by users; the main plotting method is exposed dynamically by the attributes
trueq.CircuitCollection.plotandtrueq.estimate.EstimateCollection.plotif relevant.- static plot(fit, observables=None, amplifications=None, show_imaginary=True, n_columns=3, axes=None, filename=None, legend=True)
Plots a summary of the NOX data for each applicable estimate, displaying subplots that show diagnostic information of the individual NOX circuits.
These plots can quickly tell you, e.g., if the number of NOX circuits is too small to guarantee the desired levels of statistical accuracy.
import trueq as tq from trueq.simulation import GateMatch # a simple circuit circuit = tq.Circuit() circuit += {(0, 1): tq.Gate.cx, (2, 3): tq.Gate.cx} circuit += {(1, 2): tq.Gate.cx} circuit += {(0, 1): tq.Gate.cx, (2, 3): tq.Gate.cx} circuit.measure_all() # generate the nox circuits circuits = tq.make_nox(circuit) # instantiate a simulator sim = tq.Simulator().add_depolarizing(0.02, match=GateMatch(tq.Gate.cx)) sim.run(circuits, n_shots=10000) # fit the data labels = ((0, 1, 2), (1, 2, 3)) observables = ["000", "111"] fit = circuits.fit(labels=labels, observables=observables) # plot the estimates fit.plot.raw_nox(observables=["000"])
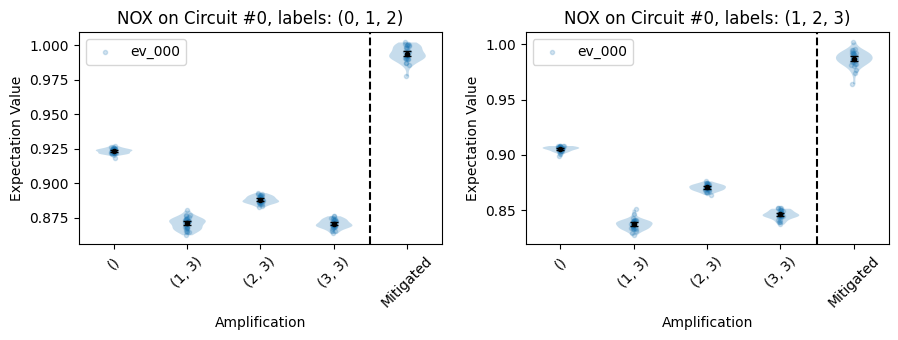
- Parameters:
fit (
EstimateCollection|CircuitCollection) – A collection of circuits or their fit.observables (
list|NoneType) – A list of strings representing observables specifying which expectation values to plot, e.g.'000','ZIZ'or'W01W02W00'. Ifobservables == None, the plotter shows the expectation value of every observable infit. Otherwise, the plotter shows the expectation value of every observable inobservablesthat is also infit. Default isNone.amplifications (
list) – A list of tuples(marker, rep), wheremarkeris the marker of a cycle being replaced by identity insertion andrepis the number of cycles that replace it. Ifamplifications == None, the plotter shows the expectation values of every NOX circuit, as well as the mitigated estimates. Otherwise, it shows the expectation values of each NOX circuit whosekey.amplificationis contained inamplifications, alongside the mitigated expecation values. Default isNone.show_imaginary (
bool) – Whether to plot the imaginary components of complex expectation values. Default is True.n_columns (
int) – If new axes are made, how many columns in the grid to use.axes (
Iterable) – An iterable of existing axes. If none are provided, new axes are created. Note that axis instances can be repeated in this list if you want to force the combination of subfigures that would normally be distinct, seeaxes()for an example.filename (
str) – An optional filename to save the file as. The extension determines the filetype; common choices are “.png”, “.pdf”, and “.svg”. If no extension is given, PNG is chosen.legend (
bool) – Whether or not the legend should be plotted. Default is True.
KNR Heatmap
- class trueq.visualization.plotters.nr.knr_heatmap
Plots KNR data as a heatmap.
Note
This class typically should not be invoked directly by users; the main plotting method is exposed dynamically by the attributes
trueq.CircuitCollection.plotandtrueq.estimate.EstimateCollection.plotif relevant.- static plot(fit_or_circuits, graph=None, width=None, cutoff=0.2, cutoff_relative_to_max=True, cutoff_ci=0.95, common_ylim=True, single_colorbar=True)
Plots all KNR data given in a single figure divided by whitespace into rows and columns of subplots. Each column of subplots corresponds to a unique cycle that was diagnosed with KNR, and each row of subplots corresponds to a different weight of error modes. Y-axis labels indicate which error occurred, and x-axis labels indicate which part of the cycle the error occurred on.
All reported errors are marginal errors. For example, if for a cycle
{0: tq.Gate.id, 1: tq.Gate.id, (2,3): tq.Gate.cnot}it is reported that \(Z\) error occurs on1: Iwith probability 0.1, then this is the sum of the probabilities of all length-4 Pauli errors with a \(Z\) on qubit 1. This particular error could in fact be part of a \(Z\otimes Z\) error on qubits 0 and 1 in the context of this cycle; you should look for this term in the heatmap to check.Some Pauli errors cannot be distinguished from each other using the KNR protocol depending on the symmetries of a particular gate. Such indistinguishable errors are grouped together with commas in the y-labels. For example, \(IZ\) cannot be distinguished from \(ZZ\) in a CNOT gate.
- Parameters:
fit_or_circuits (
CircuitCollection|EstimateCollection) – A circuit collection with KNR data, or an estimate collection with KNR fits.graph (
Graph) – AGraphobject to display device topologies at the top of columns.width (
float|NoneType) – The width of the figure in inches, orNonefor automatic.cutoff (
float) – A value below which data will not be displayed; a row of the plot will be hidden if every probability in the row is below the cutoff. This cutoff may be given in relative or absolute units, seecutoff_relative_to_max.cutoff_relative_to_max (
bool) – Whethercutoffis a fraction of the maximum error probability in the entire dataset, or whether it is absolute.cutoff_ci (
float) – A value of0.95here indicates that a probability value is considered below cutoff if its estimator is below the cutoff with 95% confidence.common_ylim (
float|bool) – A common maximum value for every color scale, orTrueto pick the maximum over all subplots, orFalseto have a different color scale for every subplot row.single_colorbar (
bool) – Ifcommon_ylimis notFalse, whether to draw a single color bar, or one identical colorbar for each subplot.
Raw
- class trueq.visualization.plotters.raw.raw
Very generic plotter that overlays exponential fits onto lightly processed data.
Note
This class typically should not be invoked directly by users; the main plotting method is exposed dynamically by the attributes
trueq.CircuitCollection.plotandtrueq.estimate.EstimateCollection.plotif relevant.- static plot(fit, labels=None, show_imaginary=True, n_columns=3, axes=None, filename=None, legend=True)
Plots a summary of raw data for each applicable circuit collection. Also fits all applicable data to exponential curves and displays this too.
These plots are diagnostic tools which can quickly tell you if your sequence lengths are way off, or if there has been some systematic error in data entry or circuit conversion to your format.
import trueq as tq circuits = tq.make_srb([0, 1], range(5, 50, 5), 20) tq.Simulator().add_overrotation(0.08).add_depolarizing(0.02).run(circuits) # show the exponential decays circuits.plot.raw()
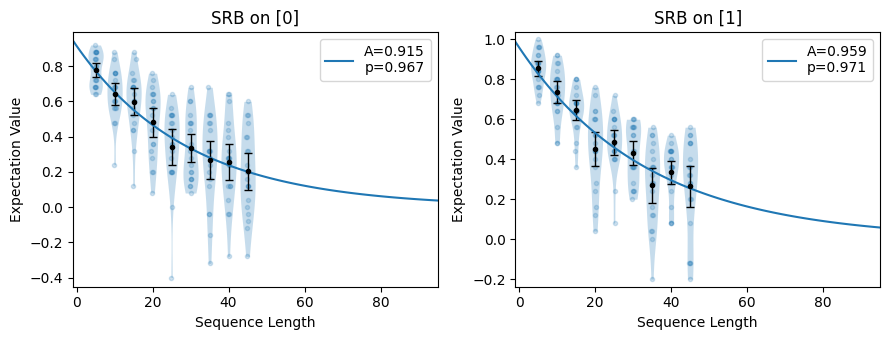
- Parameters:
fit (
EstimateCollection|CircuitCollection) – A collection of circuits or their fit.labels (
Iterable) – A list of labels (or list of lists of labels) specifying which qubit subset (or list of qubit subsets) to marginalize over.show_imaginary (
bool) – Whether to plot the imaginary components of complex decays.n_columns (
int) – If new axes are made, how many columns in the grid to use.axes (
Iterable) – An iterable of existing axes. If none are provided, new axes are created. Note that axis instances can be repeated in this list if you want to force the combination of subfigures that would normally be distinct, seeaxes()for an example.filename (
str) – An optional filename to save the file as. The extension determines the filetype; common choices are “.png”, “.pdf”, and “.svg”. If no extension is given, PNG is chosen.legend (
bool) – Whether or not the legend should be plotted. Default is True.
PlottingSuite
- class trueq.visualization.plotters.PlottingSuite(parent=None)
A collection of
Plotters, which are accessible as methods of instances of this class. Which plotters are present in the namespace is dependent on the parent object passed in at initialization. For example, a plotting function to do with XRB will only be present if the given parent has data for XRB.- Parameters:
parent (
EstimateCollection|CircuitCollection) – A circuit collection or estimate collection to store plotters for.
Plotter
- class trueq.visualization.plotters.abstract.Plotter
Represents a function that performs a plotting operation either on a
EstimateCollectionor aCircuitCollection(with data).- abstract static applies_to_estimates(keys)
A fast check that determines whether these estimates are broadly compatible with this plotting function. The plotting function is still allowed to raise (and indeed, it is encouraged) if it sees something it doesn’t like; this method is an a priori check that determines which plotters are automatically recommended to users.
- Parameters:
keys (
KeySet) – Keys of theEstimateCollection.- Return type:
bool
- abstract static applies_to_circuits(keys)
A fast check that determines whether these circuits are broadly compatible with this plotting function. The plotting function is still allowed to raise (and indeed, it is encouraged) if it sees something it doesn’t like; this method is an a priori check that determines which plotters are automatically recommended to users.
This method should usually check to see if at least some data is present.
- Parameters:
keys (
KeySet) – Keys of theEstimateCollection.- Return type:
bool
- abstract static plot()
Plots the given
EstimateCollectionorCircuitCollection.
- static axes(n_plots, axes=None, n_columns=1, axis_size=(4, 3.5), gs_options=None)
Returns an iterable of at least
n_plotsmatplotlib axes. If axes are given, those axes are returned (and repeated if there are not enough of them). If no axes are given, new axes on a grid are created using given estimates.import trueq as tq import matplotlib.pyplot as plt circuits = tq.make_srb([0, 1, 2, 3], [4, 100]) tq.Simulator().add_depolarizing(0.001).run(circuits) # plot all decays on one axis fig = plt.figure() circuits.plot.raw(axes=plt.gca()) # plot all decays on two axes in a particular order fig = plt.figure(figsize=(12, 4)) ax1, ax2 = plt.subplot(1, 2, 1), plt.subplot(1, 2, 2) circuits.plot.raw(axes=[ax1, ax2, ax2, ax1])
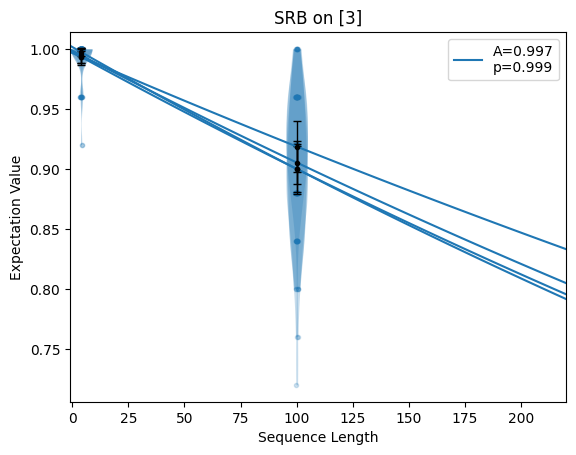
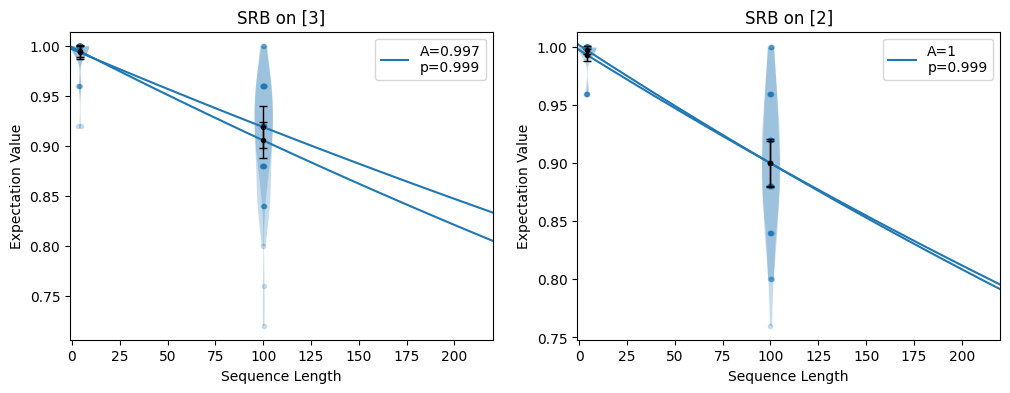
- Parameters:
n_plots (
int) – How many subfigures there will nominally be.axes (
Iterable) – An iterable of existing axes. If none are provided, new axes are created. Note that axis instances can be repeated in this list if you want to force the combination of subfigures that would normally be distinct.n_columns (
int) – If new axes are made, how many columns in the grid to use.axis_size (
tuple) – A shape tuple to use to determine the size of each subfigure.gs_options (
dict) – Options to pass tomatplotlib.gridspec.GridSpec.
- Returns:
A tuple
(axiter, finalize)whereaxiteris an iterator over axes, andfinalizeis a method to be called before the plotting function returns (this does file saving and layout tightening).- Return type:
tuple
- static savefig(filename, fig=None)
Saves the given or current figure to disk as the given filename. Saves as a PNG if no filename is given.
- Parameters:
filename (
str) – The filename to save as.fig (
matplotlib.Figure|NoneType) – A matplotlib figure to save, orNoneto use the current figure.
Timestamps
- class trueq.visualization.plotters.timestamps.timestamps
Plots the timestamps of circuit results.
Note
This class typically should not be invoked directly by users; the main plotting method is exposed dynamically by the attributes
trueq.CircuitCollection.plotandtrueq.estimate.EstimateCollection.plotif relevant.- static plot(circuits, timezone=None, axis=None, filename=None, **filter)
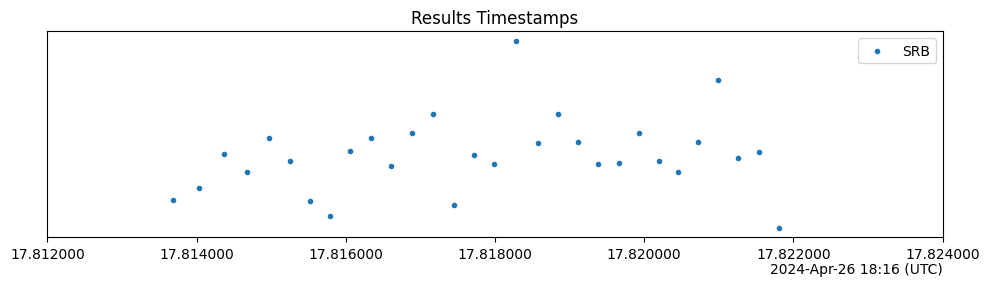
Plots the timestamp of each result in the given circuit collection, sorted by protocol type. These timestamps are taken from the
Results.last_modifiedattribute of each circuit with existing results—it is assumed that these values are parsable bydateutil.parser.parse.The y-axis is meaningless; values are chosen randomly to distinguish circuits with nearly equal timestamps.
import trueq as tq # make 30 circuits circuits = tq.make_srb(0, 5, 30) # this simulator updates the results attribute of each circuit in turn, and # new Results objects get timestamps on instantiation tq.Simulator().run(circuits) circuits.plot.timestamps()
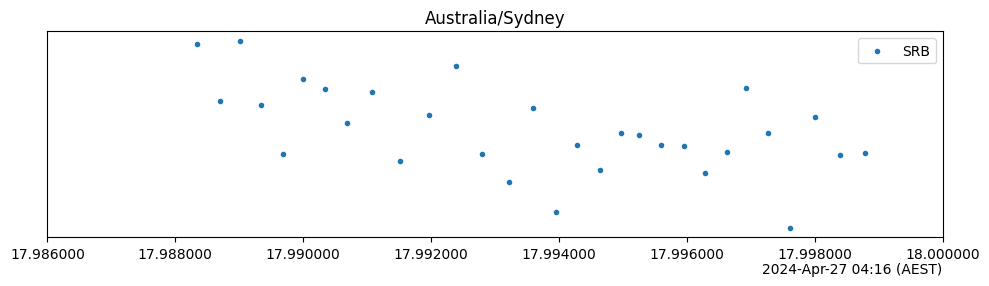
The timezone displayed on the x-axis is set to your local timezone by default, but can be changed to any timezone by providing the
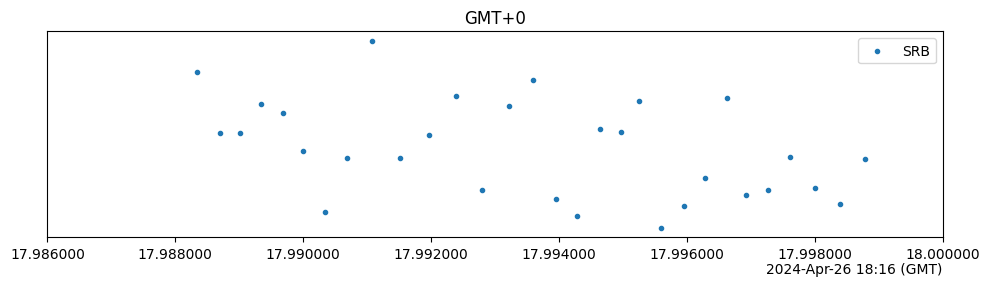
timezoneargument. The displayed timezone is completely independent of any timezone information present in the result timestamps—the relevant conversions takes place automatically. If these timestamps do not contain any timezone information, then it is assumed that they represent UTC times.import trueq as tq # make 30 circuits and populate their results circuits = tq.make_srb(0, 5, 30) tq.Simulator().run(circuits) circuits.plot.timestamps(timezone="Australia/Sydney") # The pytz package is a bit more comprehensive and not OS-dependent # You can get timezones from there, too. import pytz # list of all timezones print(pytz.all_timezones[:5] + ["..."]) circuits.plot.timestamps(timezone=pytz.timezone("GMT+0"))
['Africa/Abidjan', 'Africa/Accra', 'Africa/Addis_Ababa', 'Africa/Algiers', 'Africa/Asmara', '...']
- Parameters:
circuits (
CircuitCollection) – A circuit collection.timezone (
str|datetime.tzinfo) – The timezone to use on the x-axis.axis (
matplotlib.Axis) – An existing axis to plot on.filename (
str) – An optional filename to save the file as. The extension determines the filetype; common choices are “.png”, “.pdf”, and “.svg”. If no extension is given, PNG is chosen.**filter – A filter to apply to the datastructure, for example,
circuits.plot.timestamps(protocol="SRB").
